Proteomics Olink
Discover high-sensitivity, multiplex protein analysis with Olink— optimized for biomarker discovery and multi-omics integration.
Olink leverages Proximity Extension Assay (PEA) technology to deliver high-specificity, high-sensitivity protein quantification. With the ability to measure hundreds of proteins from a single sample, it’s a powerful platform for biomarker discovery, disease profiling, and precision medicine research.

Proteins are direct indicators of biological activity and disease states, yet have traditionally been harder to access than genomic data. Macrogen bridges this gap by combining Olink protein data with genomic insights, enabling multi-omics analysis for more personalized and predictive research outcomes.
SERVICE
Olink
Using Olink panels, Macrogen supports a full workflow from large-scale screening to target identification and validation — helping researchers accelerate protein biomarker discovery with greater accuracy and scalability.
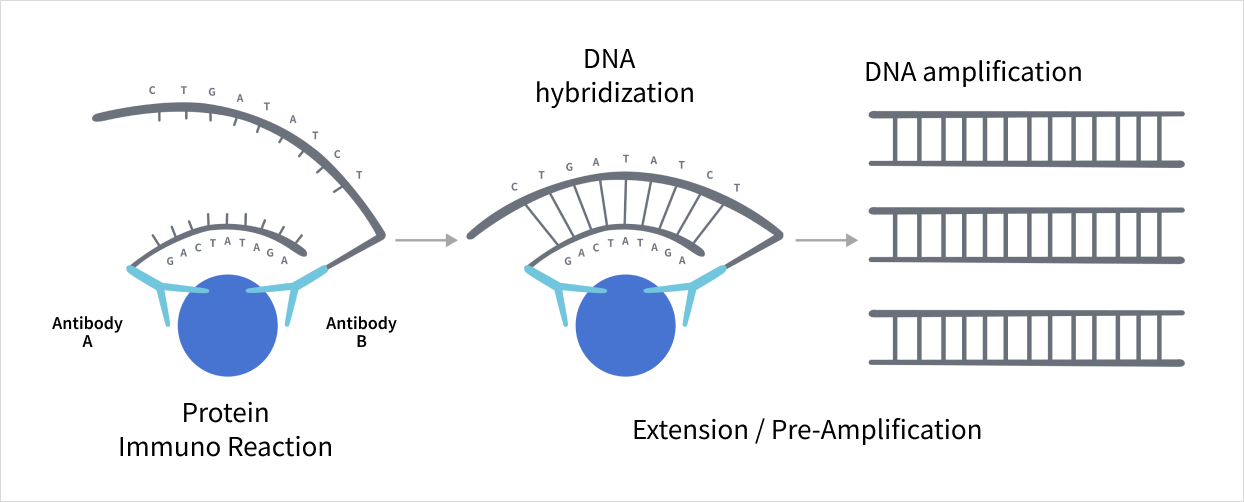
Olink’s PEA technology combines immunoassay precision with DNA-based readout. When two DNA-tagged antibodies bind to a target protein, a unique DNA sequence is generated and quantified via qPCR or NGS. This allows for precise, multiplexed measurement of protein biomarkers — even at low concentrations.
| Platform | Available Species |
No. of protein markers |
No. of sample required |
Key features | Readout Technology |
|---|---|---|---|---|---|
| Olink® Explore HT | Human | ~ 5,400 | 172 / panel |
|
NGS |
| Olink® Reveal | Human | ~ 1,000 | 86 / panel |
|
NGS |
| Olink® Target 48 | Human Mouse |
43~45 / panel | 40 / panel |
|
qPCR |
| Olink® Target 96 | Human Mouse |
92 / panel | 88 / panel |
|
qPCR |
- [Contact us for consultation]
-
- Customization options are available for Olink® Focus & Olink® Flex.
- 40 µL of serum or plasma is required. Other sample types (e.g., tissue, cells, CCM, CSF etc.) are also supported.
- If your sample quantity is below the recommended amount, please contact us for consultation.
| Areas of Analysis | Description |
|---|---|
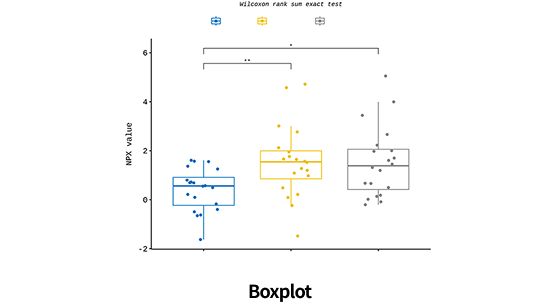
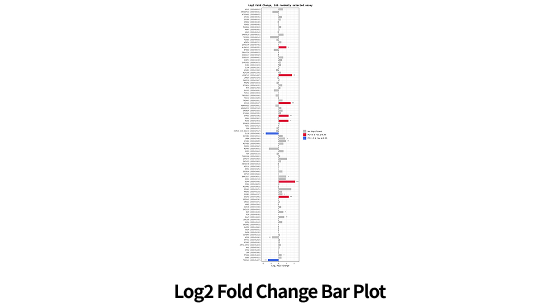
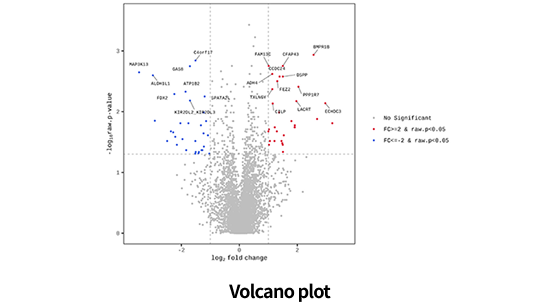
| Data analysis of DE | Basic statistics(Fold change, group mean, sd etc.) |
| Identifying differentially expressed protein(T-test, Mann-Whitney U test, etc.) | |
| Multiple testing correction (FDR, Bonferroni etc.) | |
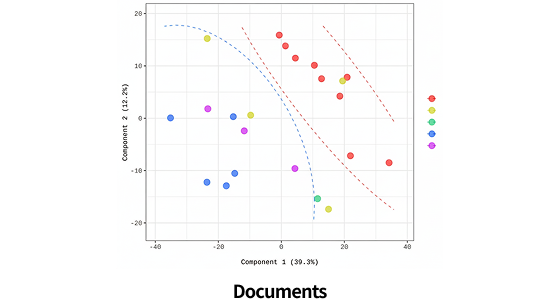
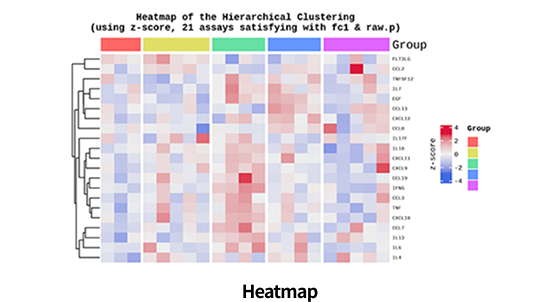
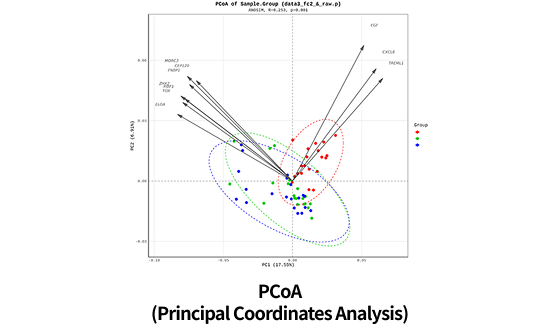
| Clustering Analysis for DEP ( Hierarchical clustering, etc.) | |
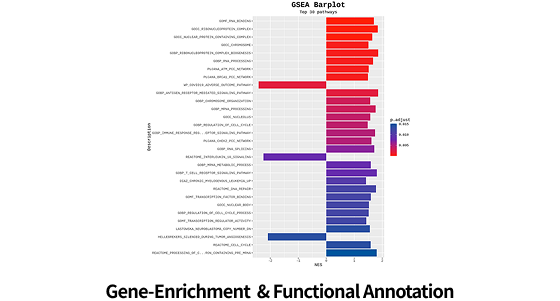
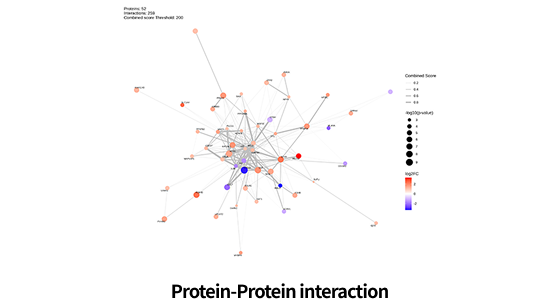
| Functional analysis |
Pathway Enrichment (Gene Set Enrichment Analysis (GSEA) / Over-representation Analysis (ORA)) |
Olink Publication
You can find a list of recent publications using Olink by visiting the link below.
Olink Publication : https://olink.com/resources-support/publications-2/