Microbiome Service
What is Metagenomics Service?
Metagenomics is a community genome analysis service that sequences the collective genomes present in various environments at once, enabling the identification of microbial diversity within those environments.
It allows the characterization of species composition and distribution within complex samples containing mixed genomes—such as feces, soil, seawater, food, as well as diverse human-derived and clinical specimens—and provides insights into their interactions and functions. This technique is widely applied to study microbial communities in different environments, focusing mainly on the diversity and distribution of bacteria, filamentous fungi, and yeasts, while also supporting eukaryotic analysis using marker gene-based approaches.
Macrogen combines advanced NGS platforms with a dedicated bioinformatics team specialized in metagenomic analysis, delivering tailored results that meet each client’s research goals.

WHAT IS MICROBIOME?
The term microbiome is a combination of “microbe” and “biome,” referring to microorganisms and their genetic information that exist in diverse environments such as the human body, soil, and the ocean.In particular, the human microbiome describes the microorganisms living within us and their genetic information, with intensive research focused on the gut microbiota.The field of studying the genomic information of these microbial communities—or the resulting data—is called metagenomics.
OUR METAGENOMICS SERVICE
Metagenomics sequences the collective genomes from various environments in a single process, enabling the identification of microbial diversity within those environments.
It characterizes species composition and distribution within complex samples containing mixed genomes—such as feces, soil, seawater, food, as well as human-derived and clinical specimens—and provides insights into their interactions and functions.This approach is widely used to analyze microbial communities, focusing on the diversity and distribution of bacteria, filamentous fungi, and yeasts, and can also be applied to eukaryotes using marker gene–based methods.At Macrogen, advanced NGS platforms and a dedicated bioinformatics team specialized in metagenomic analysis deliver customized results tailored to each client’s research needs.
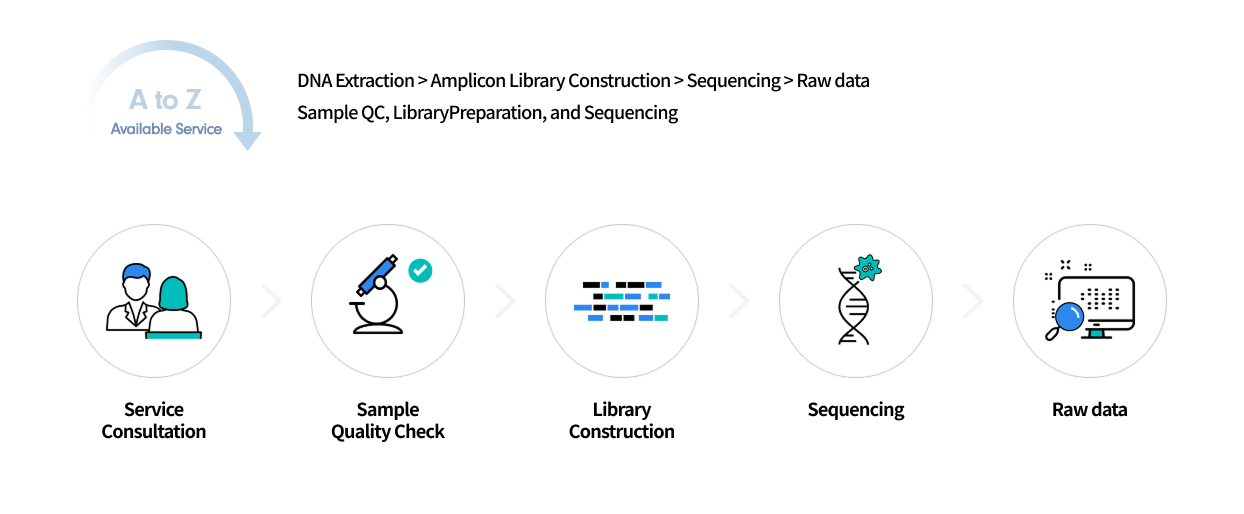
Metagenome Amplicon Sequencing
By amplifying specific regions of marker genes such as 16S rRNA or ITS, an amplicon library is constructed for sequencing-based analysis of microbial diversity and composition. This approach can also be applied to eukaryotes using the same principle of targeted marker genes.
At Macrogen, experienced bioinformatics specialists perform the analysis, ensuring highly accurate and reproducible taxonomic assignment and diversity profiling through ASV-based methods. In addition, advanced statistical and trend-based analyses are available to meet diverse research needs.
Sequencing Platform
16S rRNA V3-V4 Regions, 18S rRNA, ITS Regions, Customized regions → Illumina MiSeq i100 / Miseq
16S rRNA Full length → PacBio Revio
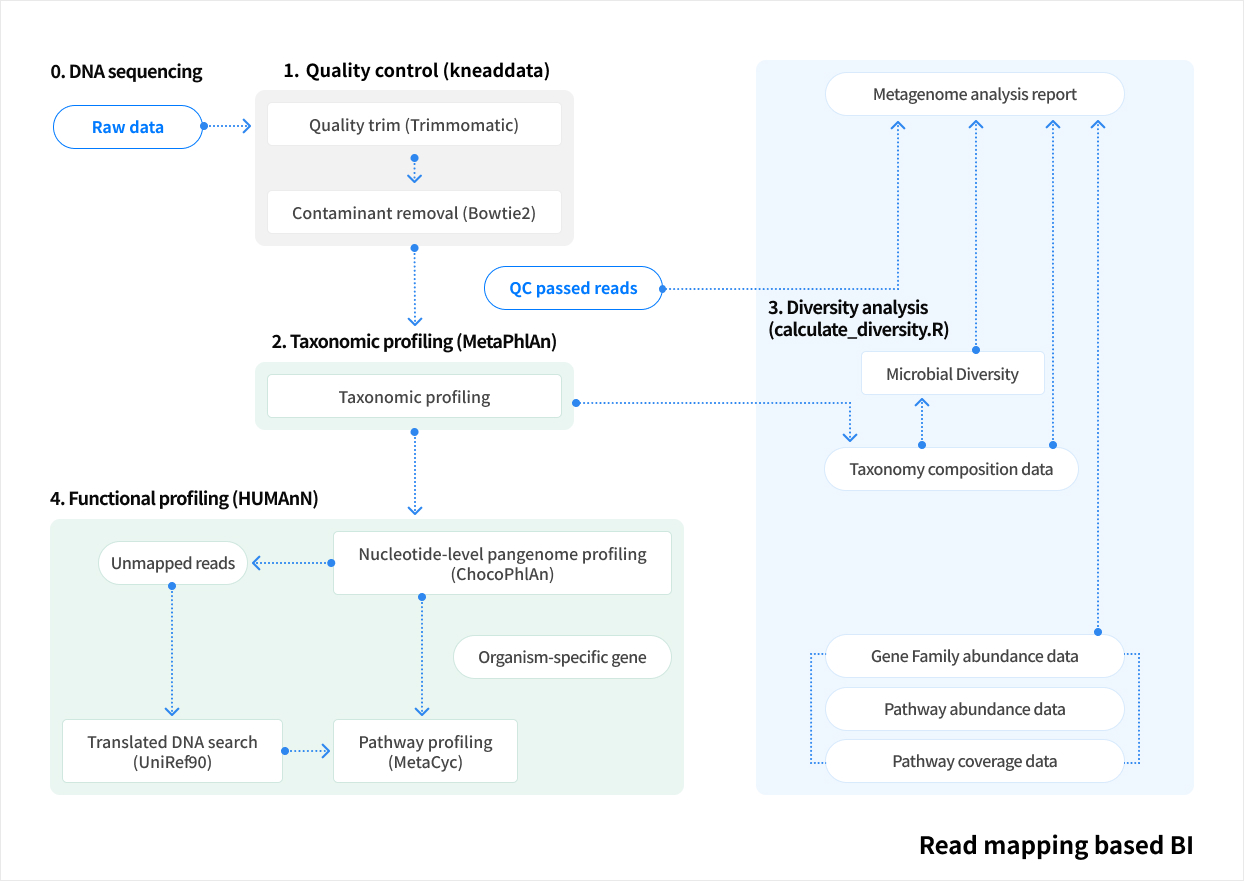
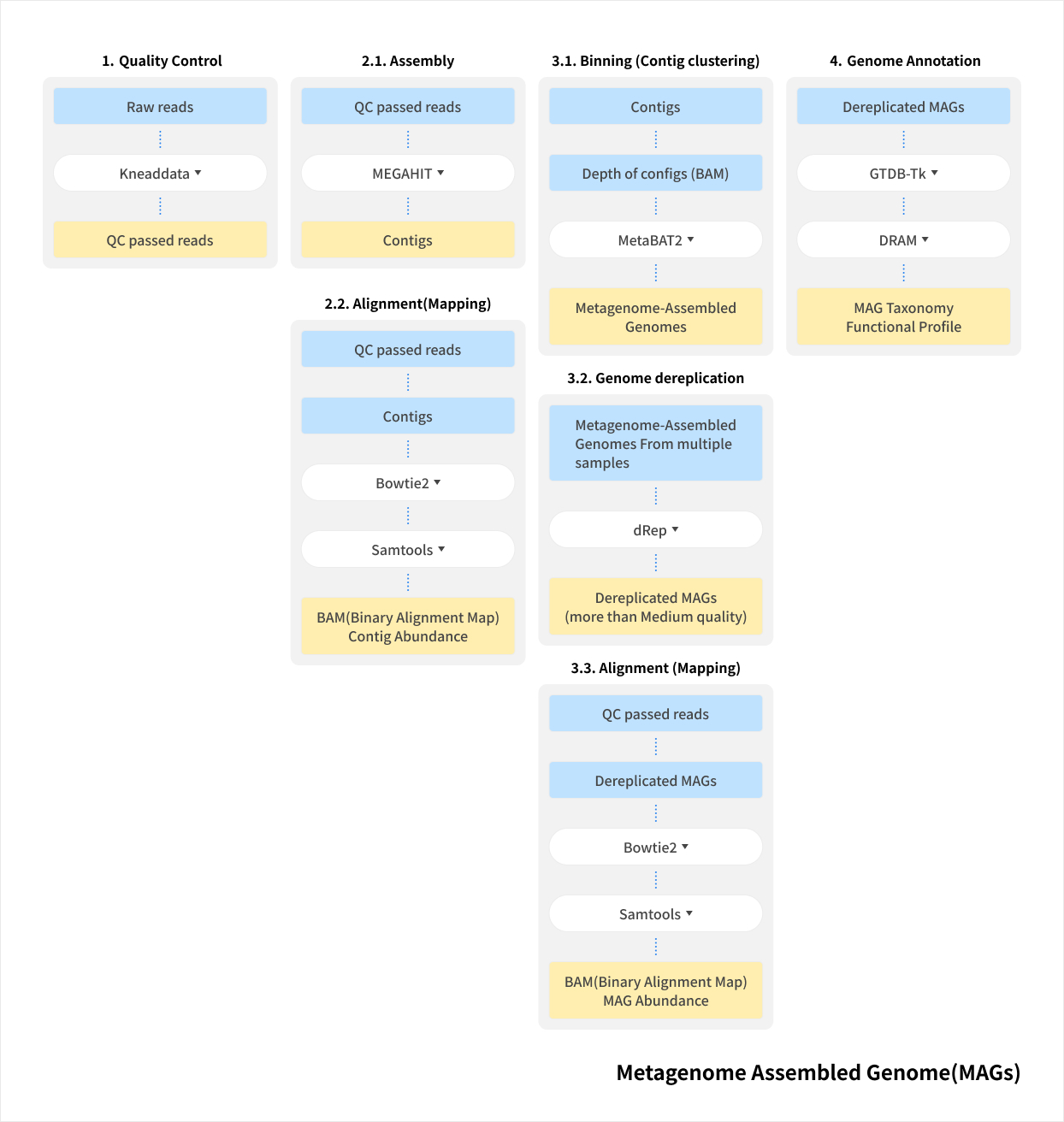
Service Process
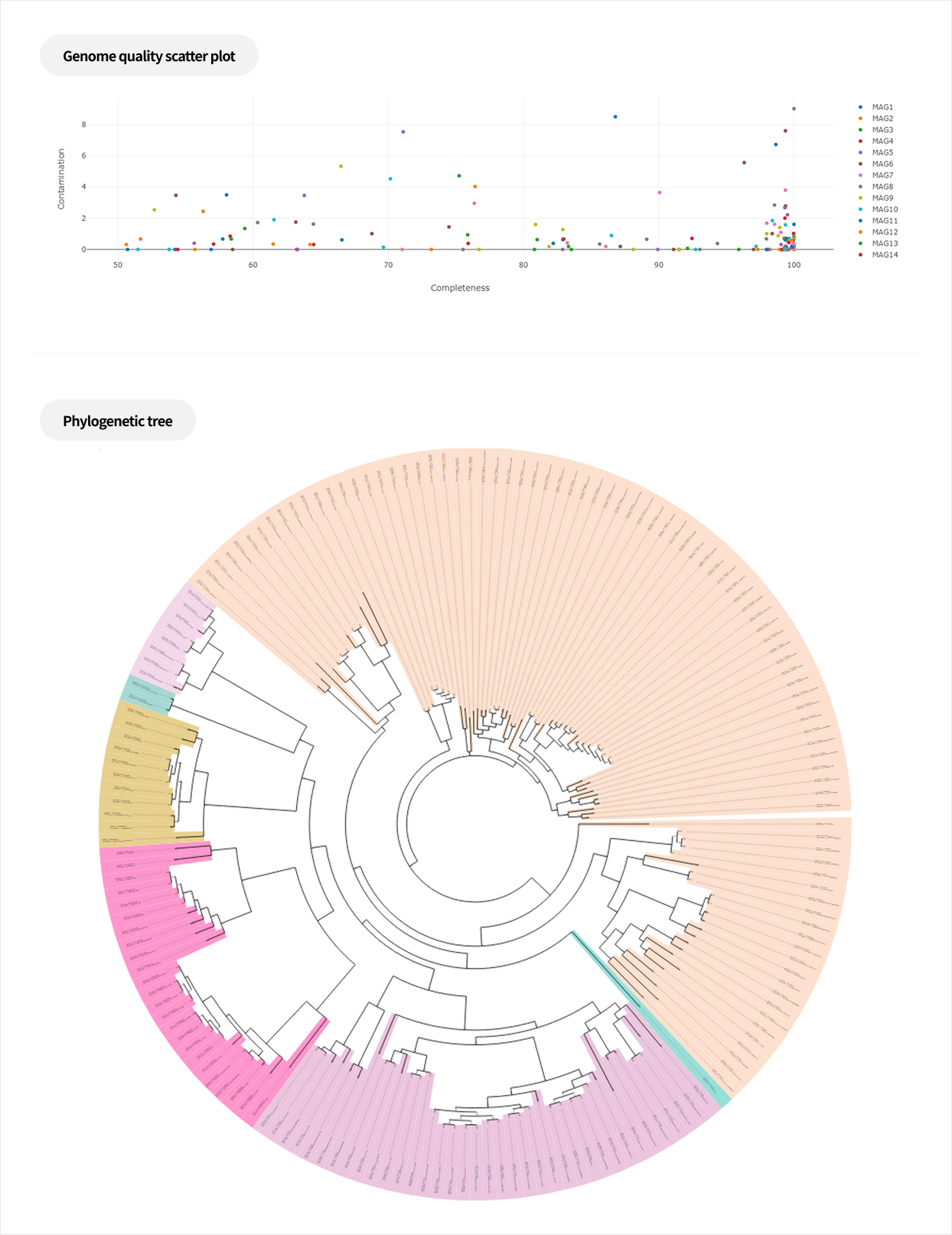
Metagenome Shotgun Sequencing
By amplifying specific regions of marker genes such as 16S rRNA or ITS, an amplicon library is constructed for sequencing-based analysis of microbial diversity and composition. This approach can also be applied to eukaryotes using the same principle of targeted marker genes.
At Macrogen, experienced bioinformatics specialists perform the analysis, ensuring highly accurate and reproducible taxonomic assignment and diversity profiling through ASV-based methods. In addition, advanced statistical and trend-based analyses are available to meet diverse research needs.
Sequencing Platform
Short read based : Illumina NovaSeq X Plus / NovaSeq 6000
Long read based : PacBio Revio

Service Process
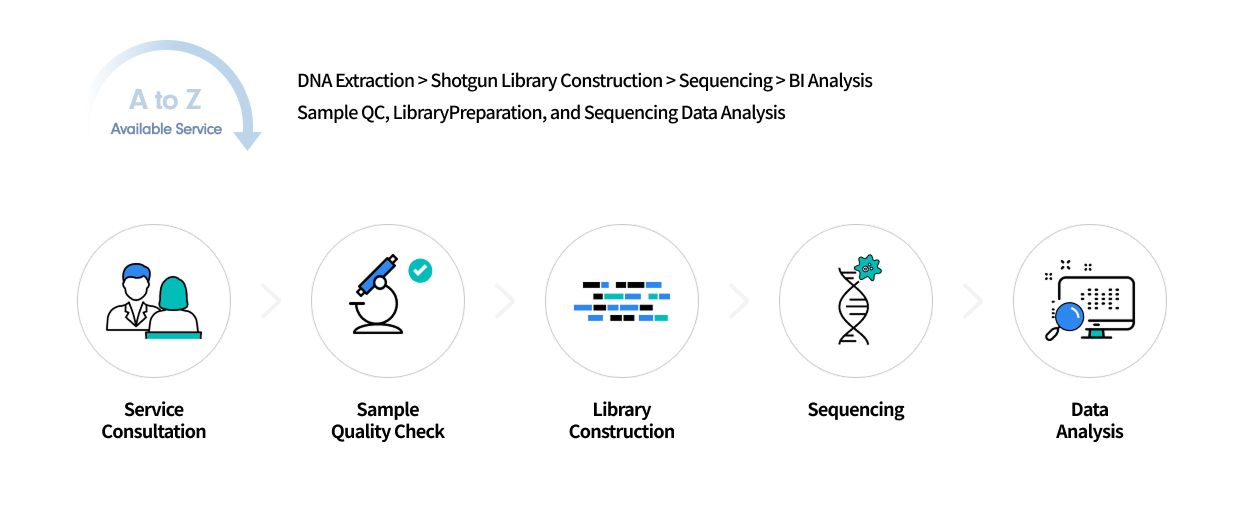
Work flow
Metagenome Transcriptome Sequencing
It enables the analysis of gene expression by extracting RNA from metagenomic samples.
Sequencing Platform
Illumina NovaSeq X Plus / NovaSeq 6000

Service Process
BI Analysis
coming soon
Customized BI
Macrogen’s metagenomics service provides a wide range of customized analysis solutions. Our dedicated team, specialized exclusively in metagenomics, offers tailored support from the research design stage through expert consultation, drawing on extensive experience with diverse sample types.
Beyond standard outputs, we provide advanced analytical options. To ensure accessibility, we also offer consultation meetings, enabling clear communication and support even for researchers with limited prior experience.
- Comprehensive Applications & Services
-
-
NGS
SERVICE -
CES
SERVICE -
DTC
SERVICE
-
- Diverse Sample Expertise
-
Advanced Expertise in
Diverse Samples
- High-Quality Sequencing
-
-
In-House
Monitoring
System -
Biannual
Internal
Reviews -
Complete
Data Spec
Oversight
-
Analysis Report
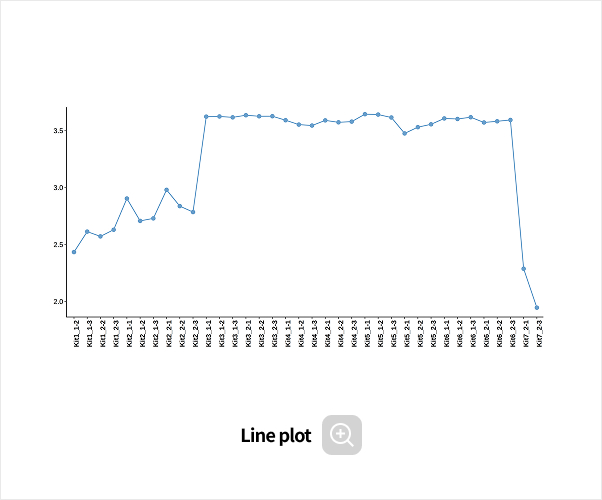
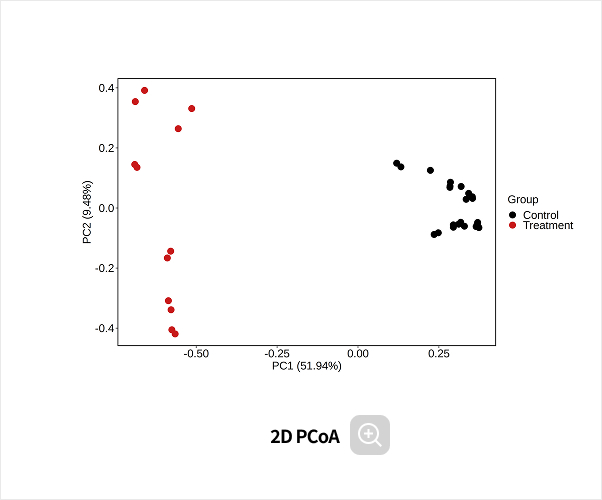
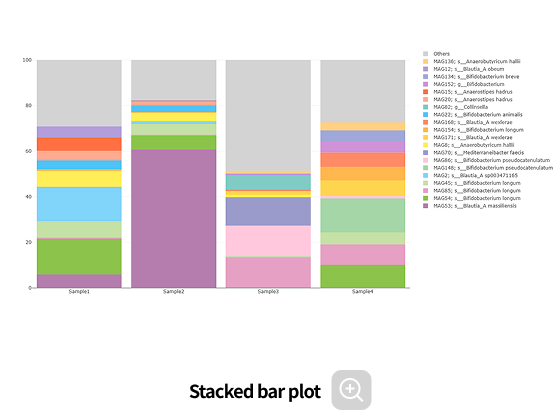
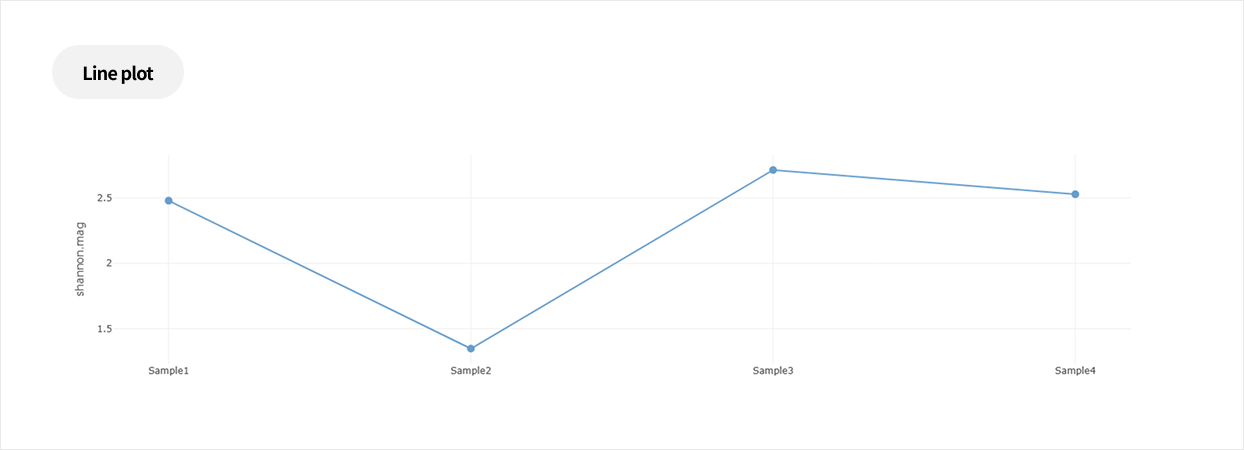
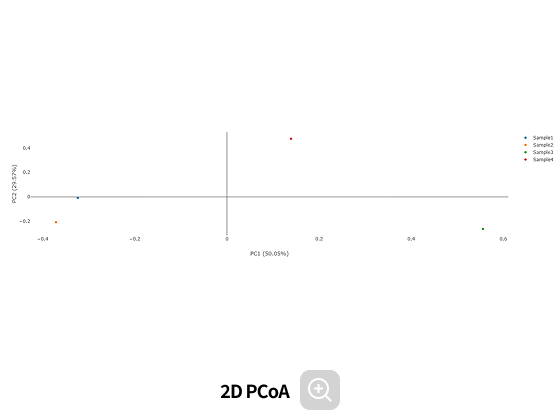
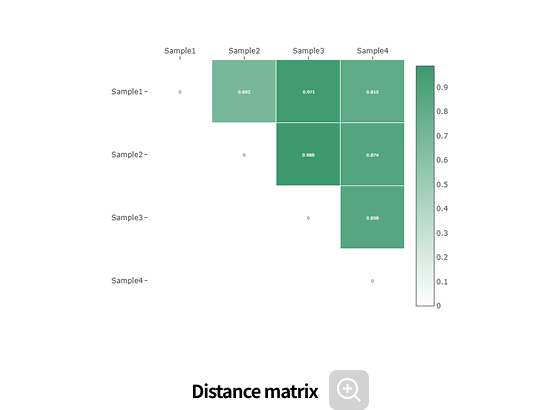
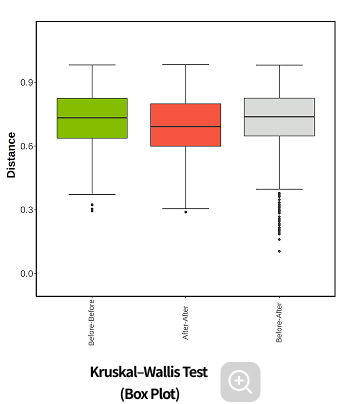
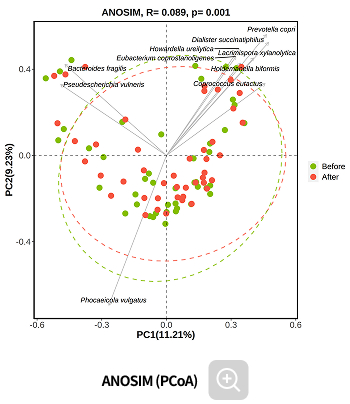
Comparative analysis - Diversity
-
This analysis compares the diversity of microbial communities across groups.
Using various diversity indices, such as α-diversity and β-diversity, it identifies intergroup differences and evaluates their statistical significance.Analysis Services- Group Statistical Analysis
(e.g., Kruskal-Wallis, Wilcoxon Test) - Intergroup Similarity Analysis
(e.g., ANOSIM, PERMANOVA)
- Group Statistical Analysis
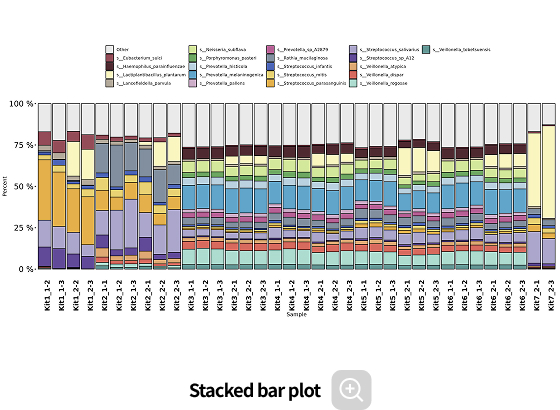
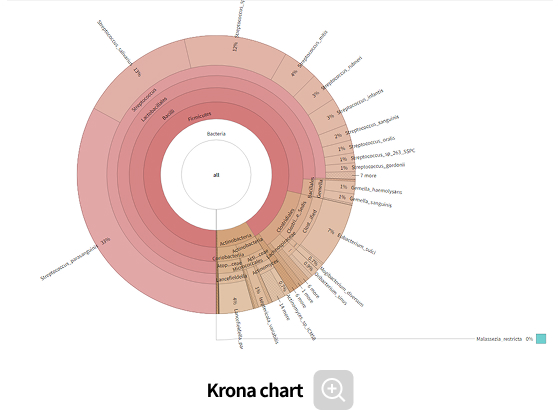
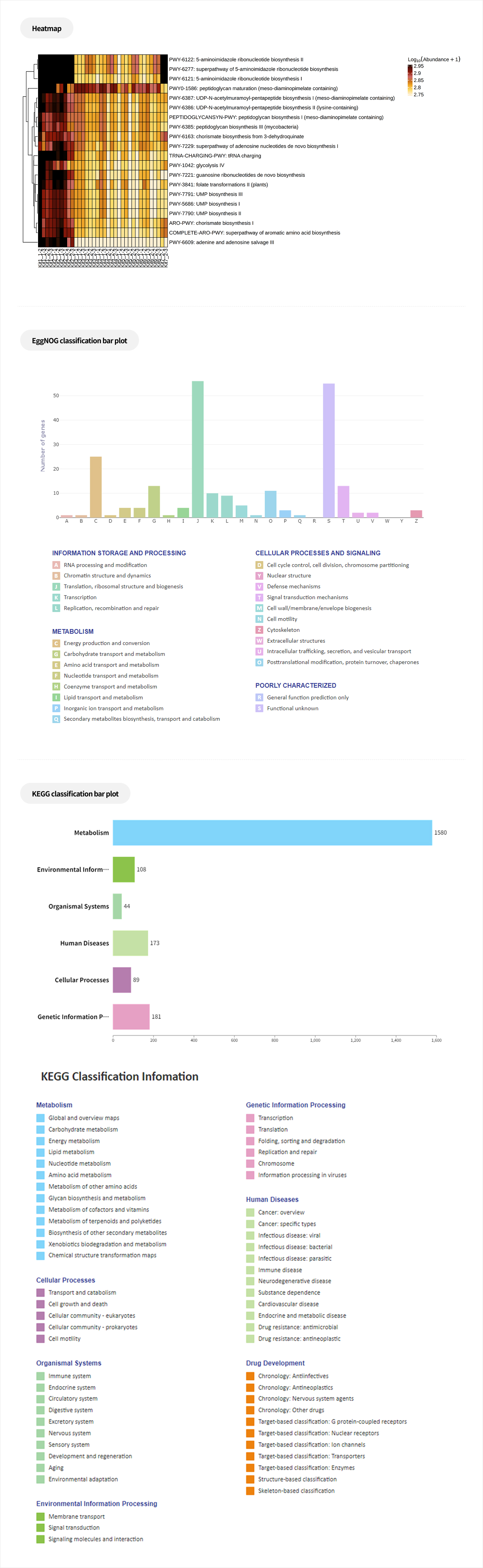
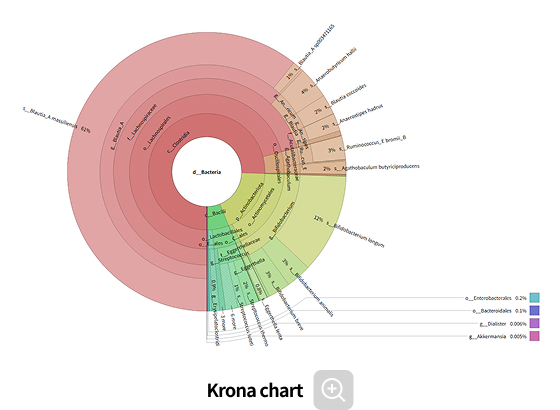
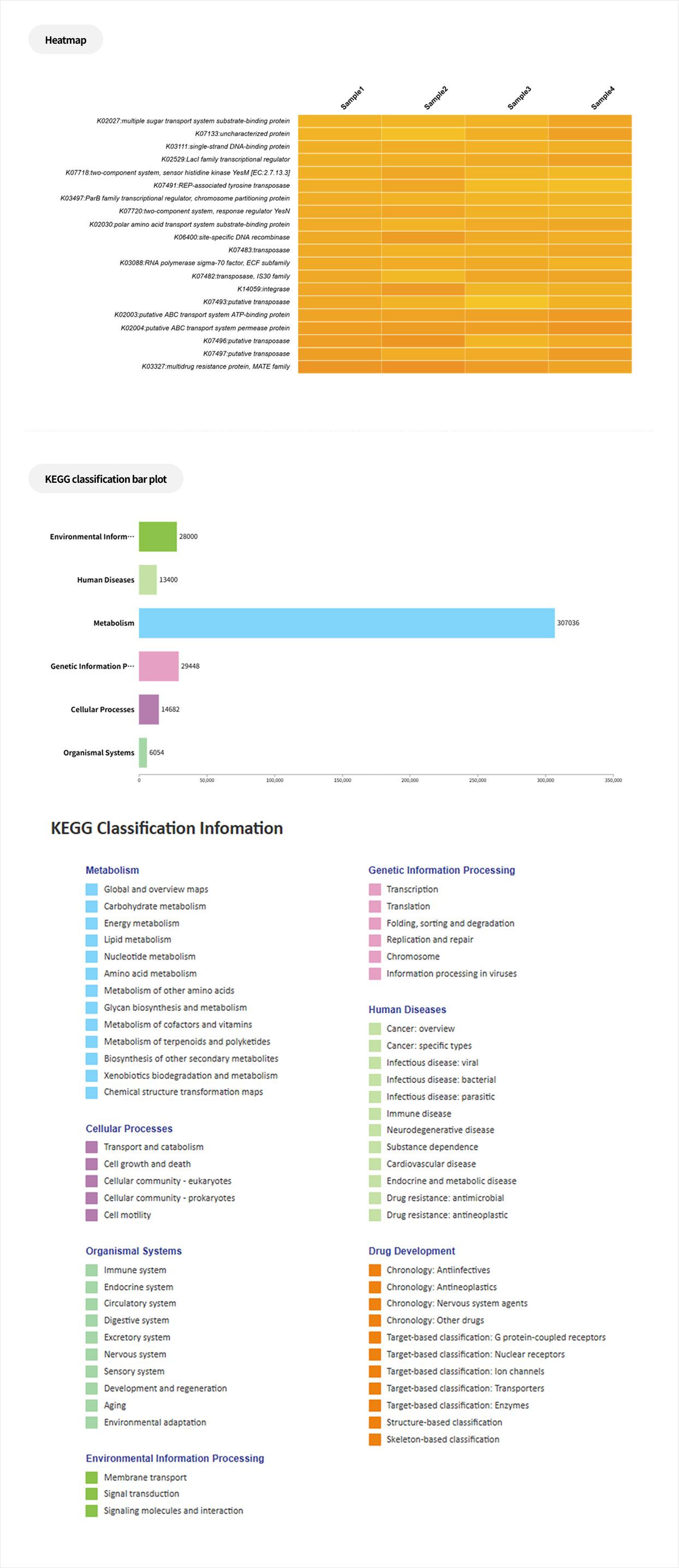
Comparative analysis - Taxonomy
-
This analysis compares microbial composition between groups, allowing identification of taxa that differ across groups. Statistical analyses and various visualization methods are employed to detect species that are specifically enriched in certain groups or exhibit significant differences in abundance.
Analysis Services- Group Statistical Analysis (e.g., Kruskal-Wallis, Wilcoxon Test)
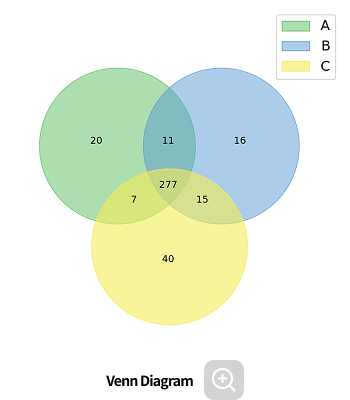
- Venn diagram
- Group bar plot, Heatmap
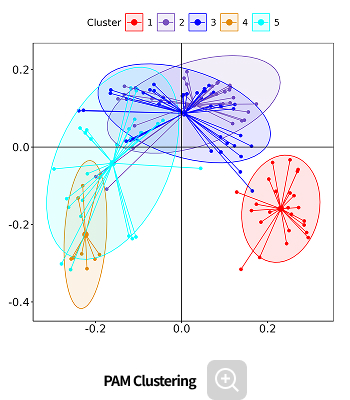
- PAM clustering
Biomarker identification
-
This analysis identifies microbial taxa that significantly contribute to differences between groups. It enables the selection of candidate microbial markers that are likely to drive intergroup variation.
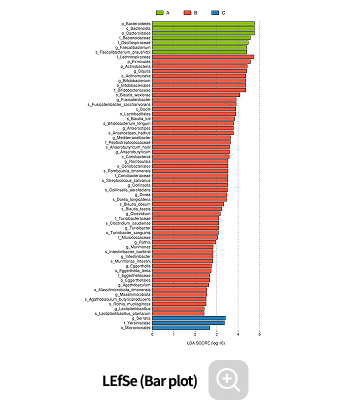
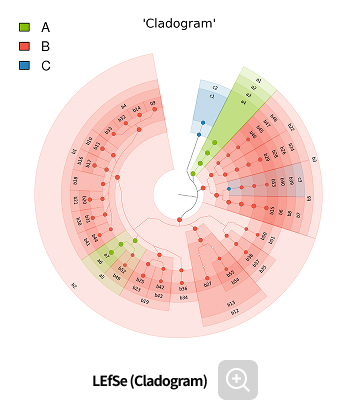
Analysis Services- LEfSe
- Random Forest
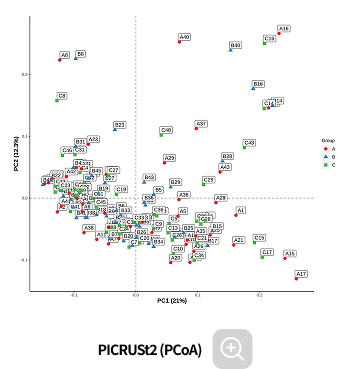
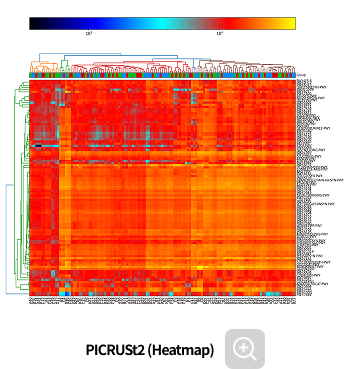
Functional prediction
-
This analysis predicts the biological functions associated with microbial communities. By integrating statistical analyses, it allows the identification of biological functions that show significant differences between groups.
Analysis Services- PIRCUSt2(MetaCyc, KEGG)
Co-occurrence analysis
-
This analysis identifies microbial modules that interact within a specific environment. It enables visualization of correlation-based networks and allows inference of community characteristics based on network properties.
Analysis Services- Correlation
- Network
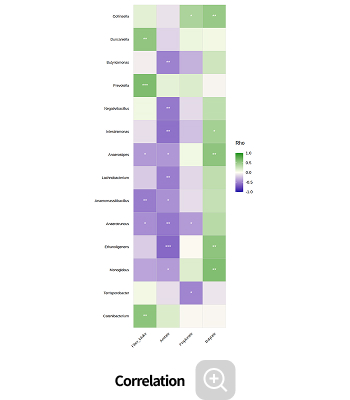
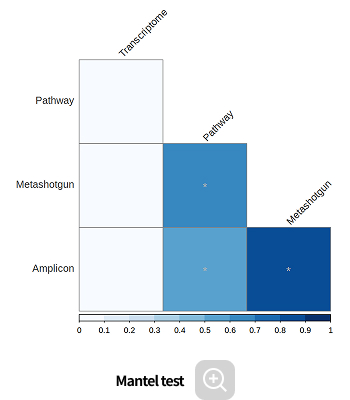
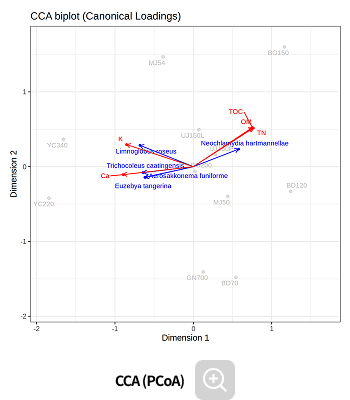
Environmental factor analysis
-
This analysis identifies microbes whose abundance is influenced by environmental factors, as well as the key environmental variables that significantly impact microbial community composition, using correlation-based methods.
Analysis Services- Correlation
- CCA
- Mantel test